Article NYT
Adaptation d'après l'article du New York Times :
Bad News Wrapped in Protein: Inside the Coronavirus Genome
https://www.nytimes.com/interactive/2020/04/03/science/coronavirus-genome-bad-news-wrapped-in-protein.html
By Jonathan Corum and Carl ZimmerApril 3, 2020
The genome of the new coronavirus is less than 30,000 “letters” long. (The human genome is over 3 billion.) Scientists have identified genes for as many as 29 proteins, which carry out a range of jobs from making copies of the coronavirus to suppressing the body’s immune responses.
The first sequence of RNA letters reads:
|
auuaaagguuuauaccuucccagguaacaaaccaaccaacuuucgaucucuuguagaucuguu
cucuaaacgaacuuuaaaaucuguguggcugucacucggcugcaugcuuagugcacucacgca
guauaauuaauaacuaauuacugucguugacaggacacgaguaacucgucuaucuucugcagg
cugcuuacgguuucguccguguugcagccgaucaucagcacaucuagguuucguccgggugug
accgaaagguaag
|
This sequence recruits machinery inside the infected cell to read the RNA letters — a, c, g and u — and translate them into coronavirus proteins.
Note: The four letters of DNA are A, C, G and T. In RNA molecules like the coronavirus genome, the T (thymine) is replaced with U (uracil).
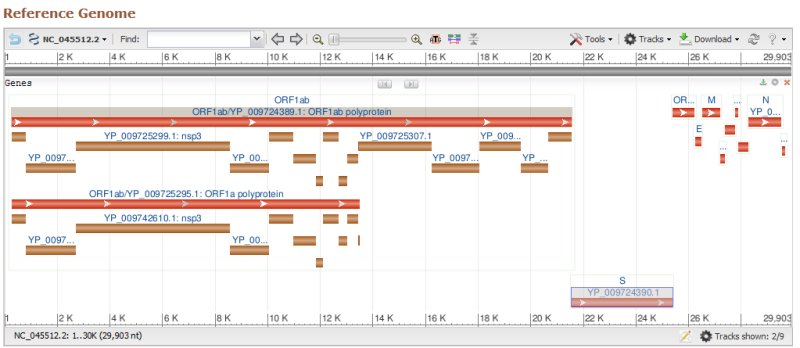
Génome du SARS-COV-2
Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome.
NCBI Reference Sequence: NC_045512.2
29903 bases en ADN.
Structure du génome de SARS-COV-2
A Chain of Proteins · ORF1ab
protein_id="YP_009724389.1"
The first viral protein created inside the infected cell is actually a chain of 16 proteins joined together. Two of these proteins act like scissors, snipping the links between the different proteins and freeing them to do their jobs.
|
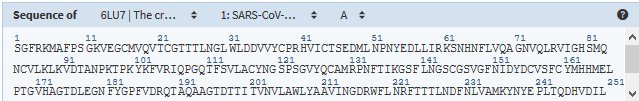
Structure en 3D sur RCSB-PDB
6LU7 : SARS-COV-2-main protease in complex with an inhibitor N3
|
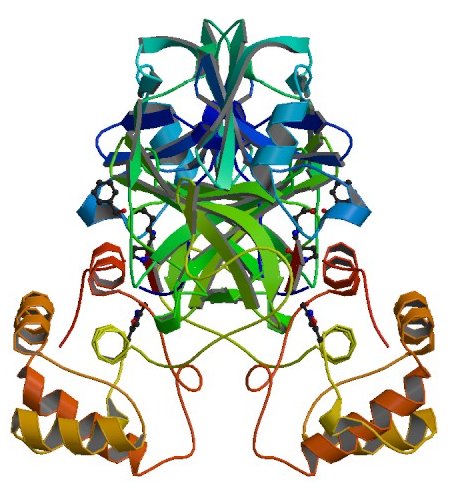
Research on other coronaviruses has given scientists a good understanding of what some of the SARS-CoV-2 proteins do. But other proteins are far more mysterious, and some might do nothing at all.
Cellular Saboteur · NSP1
protein_id="YP_009725297.1"
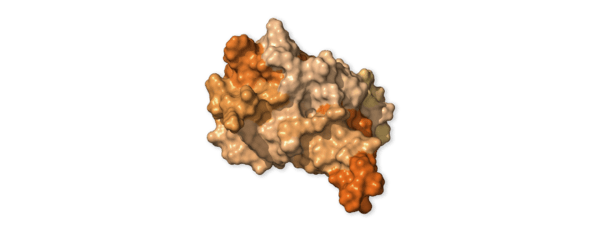
This protein slows down the infected cell’s production of its own proteins. This sabotage forces the cell to make more virus proteins and prevents it from assembling antiviral proteins that could stop the virus.
Mystery Protein · NSP2
protein_id="YP_009725298.1"
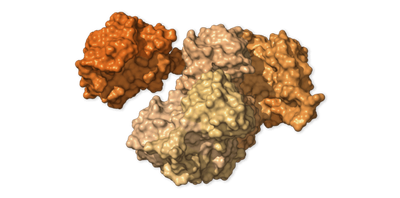
Scientists aren’t sure what NSP2 does. The other proteins it attaches to may offer some clues. Two of them help move molecule-filled bubbles called endosomes around the cell.
Untagging and Cutting · NSP3
protein_id="YP_009725299.1"
Extrait de PDB 6W6Y Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2 in complex with AMP
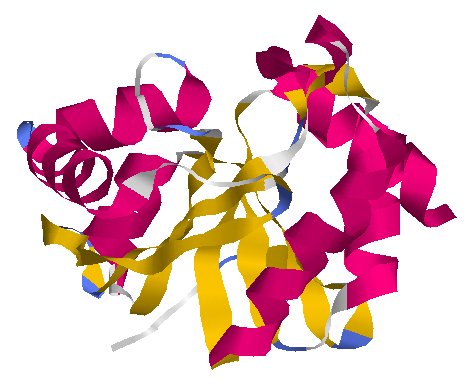
NSP3 is a large protein that has two important jobs. One is cutting loose other viral proteins so they can do their own tasks. It also alters many of the infected cell’s proteins.
Normally, a healthy cell tags old proteins for destruction. But the coronavirus can remove those tags, changing the balance of proteins and possibly reducing the cell’s ability to fight the virus.
Bubble Maker · NSP4
protein_id="YP_009725300.1"
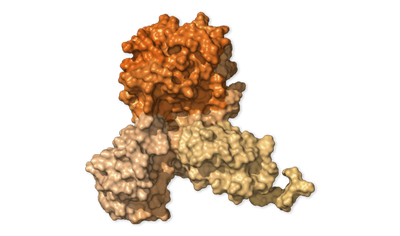
Combining with other proteins, NSP4 helps build fluid-filled bubbles within infected cells. Inside these bubbles, parts for new copies of the virus are constructed.
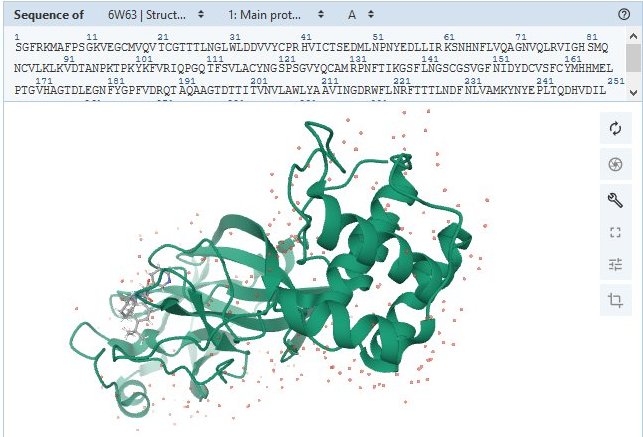
Protein Scissors · NSP5
protein_id="YP_009725301.1"
"nsp5A_3CLpro and nsp5 B_3CLpro; main proteinase (Mpro);
Structure of COVID-19 main protease bound to potent broad-spectrum non-covalent inhibitor X77
|
Structure en 3D sur RCSB-PDB : 6W63 : Structure of COVID-19 main protease bound to potent broad-spectrum non-covalent inhibitor X77
|
This protein makes most of the cuts that free other NSP proteins to carry out their own jobs.
Bubble Factory · NSP6
protein_id="YP_009725302.1"

Works with NSP3 and NSP4 to make virus factory bubbles.
Copy Assistants · NSP7 and NSP8
protein_id="YP_009725303.1"
Extrait de PDB 6M71 SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors
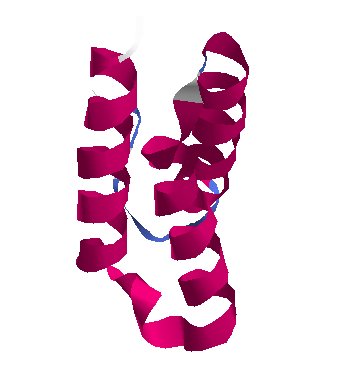
protein_id="YP_009725304.1"
Extrait de PDB 6M71 SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors
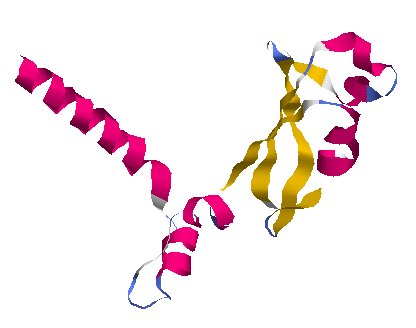
These two proteins help NSP12 make new copies of the RNA genome, which can ultimately end up inside new viruses.
At the Heart of the Cell · NSP9
protein_id="YP_009725305.1"

This protein infiltrates tiny channels in the infected cell’s nucleus, which holds our own genome. It may be able to influence the movement of molecules in and out of the nucleus — but for what purpose, no one knows.
|
Voir aussi sur RCSB-PDB 6W9Q : Peptide-bound SARS-CoV-2 Nsp9 RNA-replicase 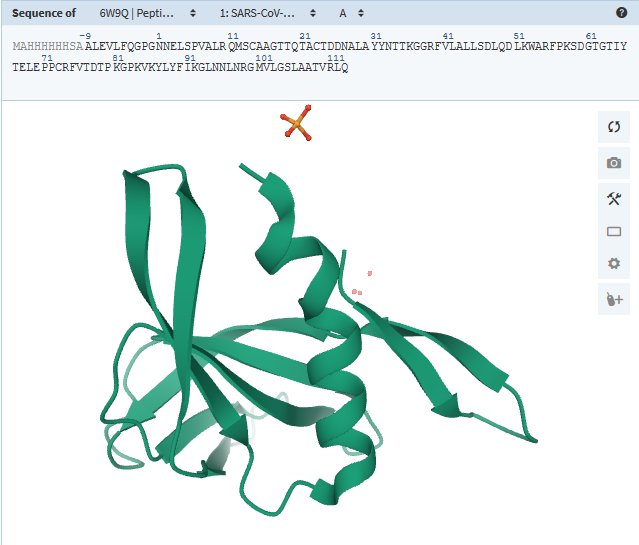
|
Genetic Camouflage · NSP10
protein_id="YP_009725306.1"
Extrait de PDB SW4H Crystal Structure of NSP16 - NSP10 Complex from SARS-CoV-2
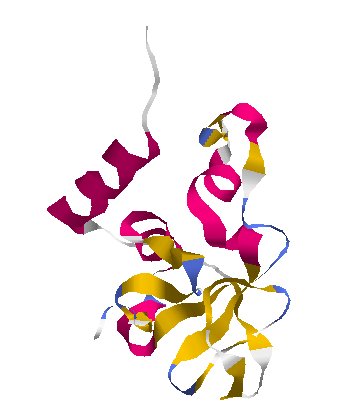
Human cells have antiviral proteins that find viral RNA and shred it. This protein works with NSP16 to camouflage the virus’s genes so that they don’t get attacked.
NSP11 et NSP12
/protein_id="YP_009725312.1"
/note="nsp12; NiRAN and RdRp;
protein_id="YP_009725307.1"
Extrait de PDB 6M71 SARS-Cov-2 RNA-dependent RNA polymerase in complex with cofactors
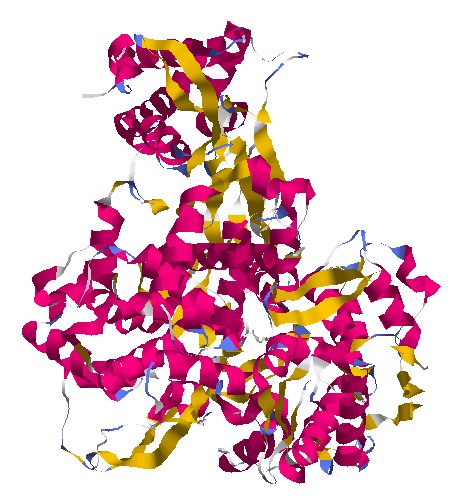
This protein assembles genetic letters into new virus genomes. Researchers have found that the antiviral remdesivir interferes with NSP12 in other coronaviruses, and trials are now underway to see if the drug can treat Covid-19.
Another sequence, NSP11, overlaps part of the same stretch of RNA. But it’s not clear if the tiny protein encoded by this gene has any function at all.
Unwinding RNA · NSP13
protein_id="YP_009725308.1"

Normally, virus RNA is wound into intricate twists and turns. Scientists suspect that NSP13 unwinds it so that other proteins can read its sequence and make new copies.
Viral Proofreader · NSP14
protein_id="YP_009725309.1"

As NSP12 duplicates the coronavirus genome, it sometimes adds a wrong letter to the new copy. NSP14 cuts out these errors, so that the correct letter can be added instead.
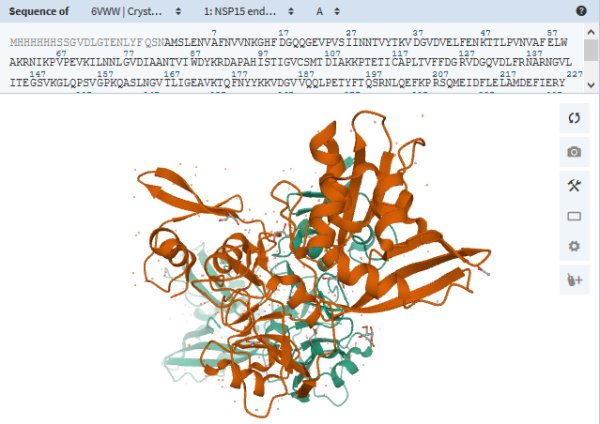
Cleaning Up · NSP15
protein_id="YP_009725310.1"
|
Structure en 3D sur RCSB-PDB : 6VWW : Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2
|
Researchers suspect that this protein chops up leftover virus RNA as a way to hide from the infected cell’s antiviral defenses.
More Camouflage · NSP16
protein_id="YP_009725311.1"
Extrait de PDB 6W4H Crystal Structure of NSP16 - NSP10 Complex from SARS-CoV-2
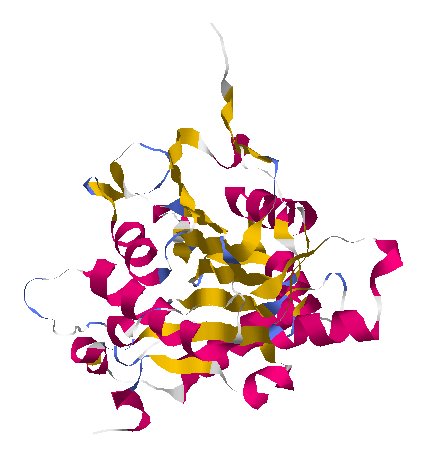
NSP16 works with NSP10 to help the virus’s genes hide from proteins that chop up viral RNA.
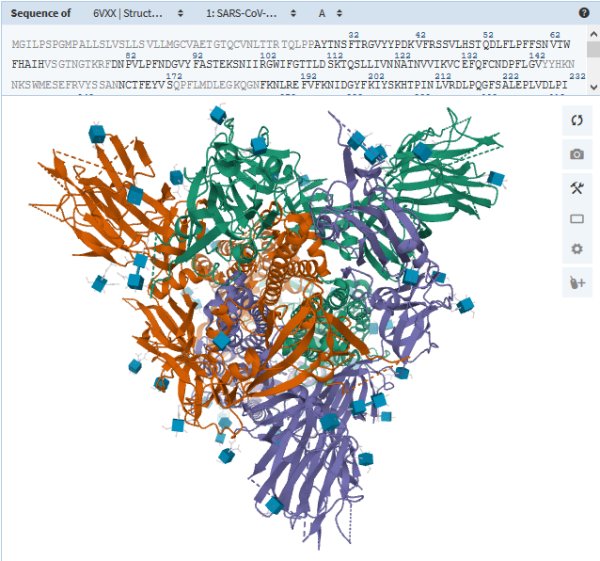
Spike Protein · S
protein_id="YP_009724390.1"

The S proteins form prominent spikes on the surface of the virus by arranging themselves in groups of three. These crownlike spikes give coronaviruses their name.
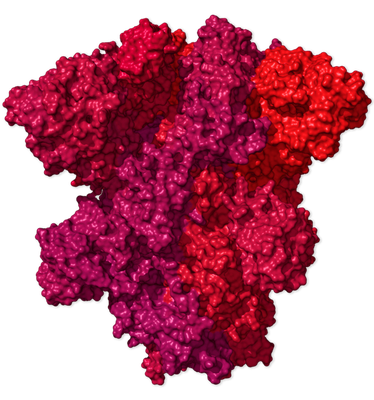
Part of the spike can extend and attach to a protein called ACE2 (in yellow below), which appears on particular cells in the human airway. The virus can then invade the cell.

The gene for the spike protein in SARS-CoV-2 has an insertion of 12 genetic letters: ccucggcgggca. This mutation may help the spikes bind tightly to human cells — a crucial step in its evolution from a virus that infected bats and other species.
|
(Voir aussi sur RCSB-PDB : 6VXX : Structure of the SARS-CoV-2 spike glycoprotein (closed state)
|
A number of scientific teams are now designing vaccines that could prevent the spikes from attaching to human cells.
Escape Artist · ORF3a
protein_id="YP_009724391.1"

The SARS-CoV-2 genome also encodes a group of so-called “accessory proteins.” They help change the environment inside the infected cell to make it easier for the virus to replicate.
The ORF3a protein pokes a hole in the membrane of an infected cell, making it easier for new viruses to escape. It also triggers inflammation, one of the most dangerous symptoms of Covid-19.
ORF3b overlaps the same RNA, but scientists aren’t sure if SARS-CoV-2 uses this gene to make proteins.
Envelope Protein · E
protein_id="YP_009724392.1"

The envelope protein is a structural protein that helps form the oily bubble of the virus. It may also have jobs to do once the virus is inside the cell. Researchers have found that it latches onto proteins that help turn our own genes on and off. It’s possible that pattern changes when the E protein interferes.
Membrane Protein · M
protein_id="YP_009724393.1"

Another structural protein that forms part of the outer coat of the virus.
Signal Blocker · ORF6
protein_id="YP_009724394.1"

This accessory protein blocks signals that the infected cell would send out to the immune system. It also blocks some of the cell’s own virus-fighting proteins, the same ones targeted by other viruses such as polio and influenza.
Virus Liberator · ORF7a - ORF7b
protein_id="YP_009724395.1"
protein_id="YP_009725318.1"

When new viruses try to escape a cell, the cell can snare them with proteins called tetherin. Some research suggests that ORF7a cuts down an infected cell’s supply of tetherin, allowing more of the viruses to escape. Researchers have also found that the protein can trigger infected cells to commit suicide — which contributes to the damage Covid-19 causes to the lungs.
Mystery Protein · ORF8
protein_id="YP_009724396.1"

The gene for this accessory protein is dramatically different in SARS-CoV-2 than in other coronaviruses. Researchers are debating what it does.
Nucleocapsid Protein · N
note="ORF9; structural protein"
product="nucleocapsid phosphoprotein"
protein_id="YP_009724397.2"

The N protein protects the virus RNA, keeping it stable inside the virus. Many N proteins link together in a long spiral, wrapping and coiling the RNA.

The accessory proteins ORF9b and ORF9c overlap this same stretch of RNA. ORF9b blocks interferon, a key molecule in the defense against viruses, but it’s not clear if ORF9c is used at all.
Mystery Protein · ORF10

Close relatives of the SARS-CoV-2 virus don’t have the gene for this tiny accessory protein, so it’s hard to know what it’s for yet — or even if the virus makes proteins from it.
End of the Line
The coronavirus genome ends with a snippet of RNA that stops the cell’s protein-making machinery.
Sources: Fan Wu et al., Nature; National Center for Biotechnology Information; Dr. David Gordon, University of California, San Francisco; Dr. Matthew B. Frieman and Dr. Stuart Weston, University of Maryland School of Medicine; Dr. Pleuni Pennings, San Francisco State University; David Haussler and Jason Fernandes, U.C. Santa Cruz Genomics Institute; Journal of Virology; Annual Review of Virology.
Model sources: Coronavirus by Maria Voigt, RCSB Protein Data Bank headquartered at Rutgers University–New Brunswick; Ribosome from Heena Khatter et al., Nature; Proteins from Yang Zhang’s Research Group, University of Michigan.