Figure 5 de Morse 3CLPro
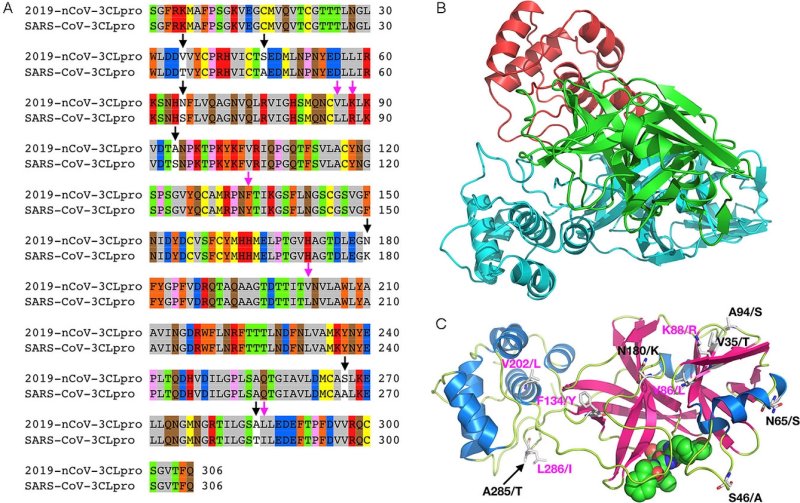
Figure 5
A) Sequence alignment for the amino acids between the 2019‐nCoV 3CLpro and the SARS‐CoV 3CLpro. Conserved (pink arrows) and nonconserved (black arrows) mutations are highlighted.
B) A 2019‐nCoV 3CLpro structure modeled by using Modeller based on the SARS‐CoV 3CLpro structure (PDB ID: 2A5I); green: catalytic domain of the first monomeric unit, red: C‐terminal domain of the first monomeric unit, cyan: second monomeric unit.
C) An alternative view of the predicted 2019‐nCoV 3CLpro monomeric structure (color‐coded by secondary structure) bound to an aza‐peptide inhibitor, showing conserved (pink) and nonconserved (black) mutations (not shown: S267/A). Residues before the forward slash refer to 2019‐nCoV; the amino acid after the slash refers to its corresponding identity in SARS‐CoV.
